|
 |
| jbm > Volume 28(3); 2021 > Article |
|
Abstract
DECLARATIONS
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (No. NRF-2019R1F1A1059208).
Authors’ contributions
Conceptualization: KWB, YKJ, and JIY; Investigation: JSP, JSK, and YSH; Writing-original draft preparation: KWB, YKJ, and SJK; Writing-review and editing: KWB, YKJ, JSP, JSK, YSH, SJK, and JIY; Project administration: JIY; Funding acquisition: JIY; All authors read and approved the final manuscript.
Fig. 1
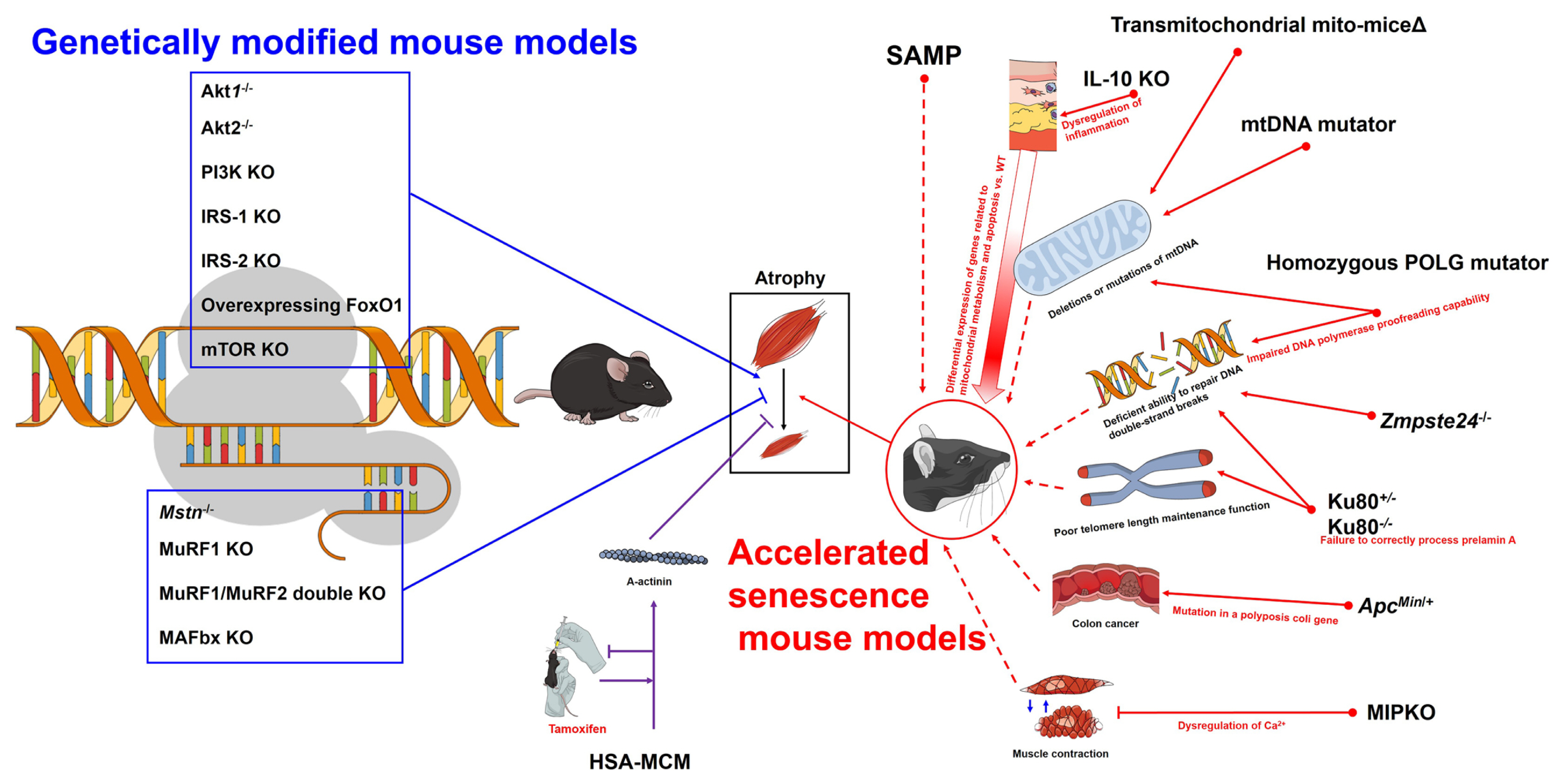
Table 1
| Model | Characteristics | Considerations | ||
|---|---|---|---|---|
| General | Detailed | |||
| SAMP | Average lifespan of less than half of normal mice (SAMP =9.7 months, SAMR=13.3 months);[5] early onset abnormal behavior, skin lesions, cataracts, elevated amyloidosis, and increased spinal lordokyphosis;[5] learning and memory deficits, degenerative joint disease, osteoporosis, tumors, and kidney dysfunction.[7] | Decreased contractility and fiber size, decreased phosphocreatine levels (SAMP8 appeared earlier than SAMP6).[4] | Type II muscle fiber size atrophy is earlier than SAMP6 (this change does not appear in EDL.[13,14] | |
|
Transmitochondrial mito-miceΔ mtDNA mutator mouse |
Premature aging phenotype, musculoskeletal disorders including sarcopenia.[16] | Muscle-specific mitochondrial aldehyde dehydrogenase 2 activity deficient mice display enhanced oxidative stress in muscle which contributes to muscle atrophy.[17] | Sarcopenia due to age-dependent accumulation of mtDNA mutations.[18] | |
| IL-10 KO mouse | Deficiency of IL-10, which regulates the immune response and acts as an anti-inflammatory agent. | Many of the genes differentially expressed from wild-type control mice are associated with mitochondrial metabolism and apoptosis.[19,20] | A good model for linking inflammation and sarcopenia. Differentiation from transmitochondrial mito-miceΔ and mtDNA mutator mice should be considered. | |
| Zmpste24−/− mouse | Mimics Hutchinson-Gilford progeria syndrome, stops growing at 5 weeks of age, and has a median survival age of 123 days.[21,22] | Loss of range of motion in the ankle after 5 weeks of age. Neuromuscular performance and selective muscle weakness.[21] | Progeria has many factors that are not suitable for expressing general aging.[23] | |
|
Ku80+/− mouse Ku80−/− mouse |
Loss of function of Ku80, which aids in repairing DNA double strand breaks and aids in telomere stability.[24] Reduction of type II muscle fiber ratio in soleus muscle (Ku80+/+ > Ku80+/− > Ku80−/−).[25] |
In the presence of normal postnatal growth, it indicates accelerated muscle aging.[24] Exhibits both reduced postnatal growth and severe progeria.[24] |
The difference in the aging phenotype between Ku80+/− and Ku80−/− in the growth process after birth should be considered. | |
| Homozygous POLG mutator mice | A homozygous knock-in mouse with impaired DNA polymerase proofreading capability.[27] | Increased mtDNA point mutations and deleted mtDNA. Reduced lifespan (median survival less than 50% of wild-type), kyphosis, osteoporosis, and alopecia. | There is some evidence for a link between mtDNA mutations and increased apoptosis,[28] but evidence for a link to oxidative stress is lacking. | |
| ApcMin/+ mouse | It is a model of colon cancer by a mutation in a polyposis coli gene,[29] and cachexia is caused by colon cancer.[30] | After 6 months of age, the weight of the gastrocnemius and soleus muscle decreases by 45% and 23%, respectively, and the average fiber cross section of the soleus muscle decreases by 24%.[31] | The muscle loss occurring in ApcMin/+ appears to be due to the high circulating levels of IL-6.[32] It is necessary to distinguished between cachexia and sarcopenia. | |
| MIPKO mouse | An animal lacking muscle-specific inositide phosphatase, an enzyme important for entry, storage, and release of Ca2+ for excitation-contraction coupling in skeletal muscle.[33] | Body weight and muscle mass loss, including reduced EDL fiber cross-sectional area, is apparent by 18 months of age compared to wild-type.[33,34] | Compared to the accelerated aging in other models, the onset of the muscle loss phenotype is relatively late. | |
| HSA-MCM mouse | Limb and craniofacial skeletal muscle-specific expression of a human α-skeletal actin protein to occur only upon the administration of a tamoxifen trigger.[35] | In genetically modified mice, incidental events may increase over time, but confusion resulting from this can be minimized. | It should be noted that tamoxifen is drug that can regulate many other hormones in the body. | |
SAMP, senescence-prone inbred strains; mtDNA, mitochondrial DNA; IL, interleukin; KO, knock-out; Zmpste24, zinc metallopeptidase STE24; POLG, DNA polymerase γ; ApcMin/+, adenomatous polyposis coli multiple intestinal neoplasia; MIPKO, muscle-specific inositide phosphatase knock-out; HSA-MCM, human skeletal muscle actin Mer-Cre-Mer; SAMR, senescence-resistant inbred strains; EDL, extensor digitorum longus.
Table 2
| Model | Characteristics | Considerations | |
|---|---|---|---|
| General | Detailed | ||
|
Akt1−/− mouse Akt2−/− mouse |
Smaller than WT littermates, but overall physiology is minimally altered, including a non-diabetic phenotype.[48] Indistinguishable in appearance at birth from WT mice but develops insulin resistance.[49] |
Both Akt1−/−, Akt2−/− showed lower mass of EDL, gastrocnemius, and quadriceps than WT.[50] | In the case of Akt2−/−, it shows an increase in the mass of the soleus muscle compared to the WT.[50] |
| PI3K KO mouse | Several PI3K KO models have exhibited normal growth, except a null model for p85α/p55α/p50α and p85β regulatory subunits that showed a reduced heart size but essentially unchanged skeletal muscle size and morphology.[51] | For, sarcopenia research, it would be good to consider using a null model for the p85α/p55α/p50α and p85β regulatory subunits. | Inhibiting of muscle development factors does not mean inducing muscle loss. Therefore, some of the PI3K mutants are not useful for studying sarcopenia. |
|
IRS-1 KO mouse IRS-2 KO mouse |
Both IRS-1 and IRS-2 contribute to muscle development, but do not have a specific phenotype for muscle physiology and provide no information on muscle loss. | There are questions about the role of IGF-1 in the area of load-induced muscle hypertrophy.[52-56] Inhibiting of muscle development factors does not mean inducing muscle loss. | |
| Mstn−/− mouse | Mstn acts as a regulator of muscle growth via activating and phosphorylating Smad2/3 transcription factors through the Activin IIB membrane receptor.[63-65] | Mstn−/− mice show increased muscle mass[64] and increased expression of anti-apoptotic factors.[69] Increased satellite cell number and type IIb/X fiber cross-sectional area in tibialis anterior muscles.[67] | Mstn−/− mice are more susceptible to atrophy caused by unloading.[70] Aged Mstn−/− mice maintain muscle mass.[72] |
| Overexpressing FoxO1 mouse | Weigh less than WT mice and have less relative muscle mass. | Compared to WT mice whose cathepsin L expression is upregulated at 3 months of age, the size of type I and type II muscle fibers is significantly reduced.[82] | The reduction in type I muscle fibers is greater than that of type II muscle fibers.[82] |
| mTOR KO mouse | Exhibits reduced weights and cross-sectional area of type II muscles, such as gastrocnemius, tibialis anterior, and plantaris, while both cross-sectional area and weight of soleus were increased (when normalized to body weight) at 6 weeks old.[84] | The morphological changes in muscle-specific mTOR KO mice are particularly pronounced in oxidative fibers, which are similar to muscular dystrophy and myopathy. Therefore, this mutation is sometimes not suitable as a model for sarcopenia study.[84] | |
|
MuRF1 KO mouse MuRF1/MuRF2 double KO mouse MAFbx KO mouse |
These are mouse transgenic mouse species that do not express well-known proteins related to muscle atrophy. |
Resisted muscle atrophy, showed about a 35% gastrocnemius muscle weight sparing after 14 days of sciatic nerve denervation injury.[85] Skeletal muscle hypertrophy and 38% higher quadriceps muscle weight compared to WT.[86] Resisted muscle atrophy, showed about a 56% gastrocnemius muscle weight sparing after 14 days of sciatic nerve denervation injury.[85] |
|
REFERENCES
- TOOLS
-
METRICS

-
- 4 Crossref
- 0 Scopus
- 6,529 View
- 210 Download
- ORCID iDs
-
Kyung-Wan Baek

https://orcid.org/0000-0002-8445-3773Youn-Kwan Jung

https://orcid.org/0000-0002-5784-6221Jin Sung Park

https://orcid.org/0000-0002-6284-9566Ji-Seok Kim

https://orcid.org/0000-0002-3023-1999Young-Sool Hah

https://orcid.org/0000-0002-8571-2722So-Jeong Kim

https://orcid.org/0000-0001-6422-7595Jun-Il Yoo

https://orcid.org/0000-0002-3575-4123 - Related articles




 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print